# Basics
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
# Probability
from scipy.stats import multivariate_normal
# Machine Learning
from sklearn.preprocessing import StandardScaler
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
from sklearn.naive_bayes import GaussianNB
# Classification Metrics
from sklearn.metrics import accuracy_score, confusion_matrix
from sklearn.metrics import ConfusionMatrixDisplayCS 307: Week 10
# Deal with pandas versus seaborn issues
warnings.simplefilter(action="ignore", category=FutureWarning)Some Bayes Theorem
Given priors:
- \(\pi_A = 0.20\)
- \(\pi_B = 0.50\)
- \(\pi_C = 0.30\)
And likelihoods:
- \(X \mid Y = A \sim N(\mu = 2, \sigma = 1)\)
- \(X \mid Y = B \sim N(\mu = 3, \sigma = 2)\)
- \(X \mid Y = C \sim N(\mu = 4, \sigma = 1)\)
Calculate posteriors:
- \(P(Y = A \mid X = 3.4)\)
- \(P(Y = B \mid X = 3.4)\)
- \(P(Y = C \mid X = 3.4)\)
import numpy as np
from scipy.stats import norm
import matplotlib.pyplot as plt
x = np.linspace(-2, 8, 1000)
mu = [2, 3, 4]
sigma = [1, 2, 1]
fig, axs = plt.subplots(1, 2, figsize=(12, 5))
for i in range(3):
y = norm.pdf(x, loc=mu[i], scale=sigma[i])
axs[0].plot(x, y, label=f"Y={chr(65+i)}", linewidth=2)
axs[0].axvline(x=3.4, color="red", linestyle="--")
axs[0].set_xlabel("X")
axs[0].set_ylabel("Density")
axs[0].grid(color="lightgrey", linestyle="--")
axs[0].set_title("Likelihoods")
axs[0].legend()
priors = [0.20, 0.50, 0.30]
z = []
for i in range(len(x)):
z.append(np.sum(priors * norm.pdf(x[i], loc=mu, scale=sigma)))
for i in range(3):
y = priors[i] * norm.pdf(x, loc=mu[i], scale=sigma[i])
axs[1].plot(x, y / z, label=f"Y={chr(65+i)}", linewidth=2)
axs[1].axvline(x=3.4, color="red", linestyle="--")
axs[1].set_xlabel("X")
axs[1].set_ylabel("Density")
axs[1].grid(color="lightgrey", linestyle="--")
axs[1].set_title("Posteriors")
axs[1].legend()
plt.show()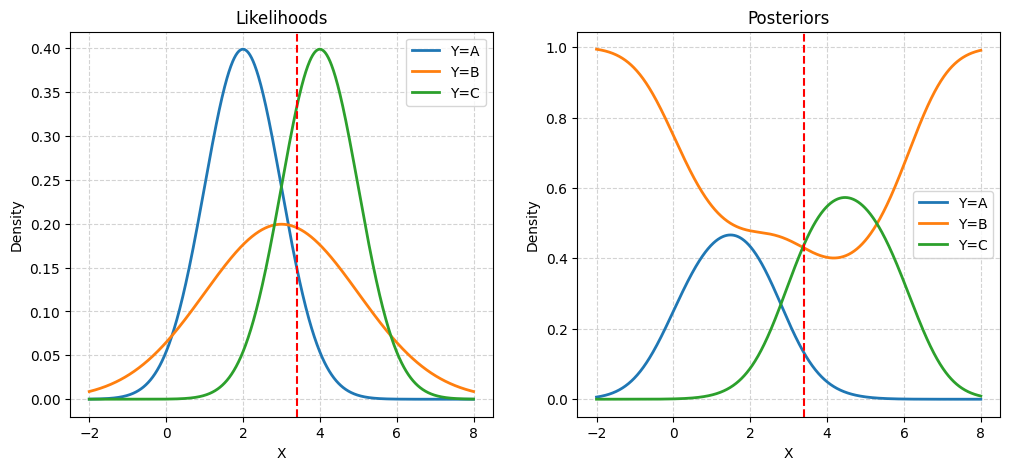
import numpy as np
from scipy.stats import norm
priors = [0.20, 0.50, 0.30]
likelihoods = norm.pdf(x=3.4, loc=[2, 3, 4], scale=[1, 2, 1])
priors * likelihoods / (np.sum(priors * likelihoods))array([0.13152821, 0.42938969, 0.43908211])- \(P(Y = A \mid X = 3.7) = 0.13152821\)
- \(P(Y = B \mid X = 3.7) = 0.42938969\)
- \(P(Y = C \mid X = 3.7) = 0.43908211\)
Generative Models
\[ p_k(\boldsymbol{x}) = P(Y = k \mid \boldsymbol{X} = \boldsymbol{x}) = \frac{\pi_k \cdot f_k(\boldsymbol{x})}{\sum_{g = 1}^{G} \pi_g \cdot f_g(\boldsymbol{x})} \]
Helper Functions
def make_plot(mu1, cov1, mu2, cov2):
# Create two multivariate normal distributions
dist1 = multivariate_normal(mu1, cov1)
dist2 = multivariate_normal(mu2, cov2)
# Generate random samples from each distribution
samples1 = dist1.rvs(size=100)
samples2 = dist2.rvs(size=100)
# Change color maps for scatter plot
cmap1 = plt.get_cmap("Blues")
cmap2 = plt.get_cmap("Oranges")
# Plot true distributions
x_min = np.min([np.min(samples1[:, 0]), np.min(samples2[:, 0])]) - 1
x_max = np.max([np.max(samples1[:, 0]), np.max(samples2[:, 0])]) + 1
y_min = np.min([np.min(samples1[:, 1]), np.min(samples2[:, 1])]) - 1
y_max = np.max([np.max(samples1[:, 1]), np.max(samples2[:, 1])]) + 1
x, y = np.mgrid[x_min:x_max:0.01, y_min:y_max:0.01]
pos = np.empty(x.shape + (2,))
pos[:, :, 0] = x
pos[:, :, 1] = y
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].contourf(x, y, dist1.pdf(pos), cmap="Blues")
ax[0].contour(x, y, dist1.pdf(pos), colors="k")
ax[0].contourf(x, y, dist2.pdf(pos), cmap="Oranges", alpha=0.5)
ax[0].contour(x, y, dist2.pdf(pos), colors="k")
ax[0].grid(color="lightgrey", linestyle="--")
ax[0].set_title("True Distribution")
ax[1].scatter(samples1[:, 0], samples1[:, 1], color=cmap1(0.5))
ax[1].scatter(samples2[:, 0], samples2[:, 1], color=cmap2(0.5))
ax[1].set_title("Sampled Data")
ax[1].grid(color="lightgrey", linestyle="--")
ax[0].set_xlim(x_min, x_max)
ax[0].set_ylim(y_min, y_max)
ax[1].set_xlim(x_min, x_max)
ax[1].set_ylim(y_min, y_max)
plt.show()# Create mean (mu) and covariance (cov) arrays
def create_params(mu_1, mu_2, sigma_1, sigma_2, corr):
# Create mean vector
mu = np.array([mu_1, mu_2])
# Calculate covariance matrix
cov = np.array([[sigma_1**2, corr * sigma_1 * sigma_2], [corr * sigma_1 * sigma_2, sigma_2**2]])
# Return both
return mu, cov# Print parameter information
def print_setup_parameter_info(mu1, cov1, mu2, cov2):
print("Mean X when Y = Blue:", mu1)
print("Covariance of X when Y = Blue:", "\n", cov1)
print("Mean X when Y = Orange:", mu2)
print("Covariance of X when Y = Orange:", "\n", cov2)Linear Discriminant Analysis (LDA)
Linear Discriminant Analysis, LDA, assumes that the features are multivariate normal conditioned on the target classes. Importantly, LDA assumes the same covariance (shape) within each class.
\[ \boldsymbol{X} \mid Y = k \sim N(\boldsymbol{\mu}_k, \boldsymbol\Sigma) \]
\[ \boldsymbol\Sigma = \boldsymbol\Sigma_1 = \boldsymbol\Sigma_2 = \cdots = \boldsymbol\Sigma_G \]
# Make a two-class LDA setup
mu1, cov1 = create_params(0, 0, 1, 1, 0.5)
mu2, cov2 = create_params(2, 2, 1, 1, 0.5)
print_setup_parameter_info(mu1, cov1, mu2, cov2)Mean X when Y = Blue: [0 0]
Covariance of X when Y = Blue:
[[1. 0.5]
[0.5 1. ]]
Mean X when Y = Orange: [2 2]
Covariance of X when Y = Orange:
[[1. 0.5]
[0.5 1. ]]# Plot it!
make_plot(mu1, cov1, mu2, cov2)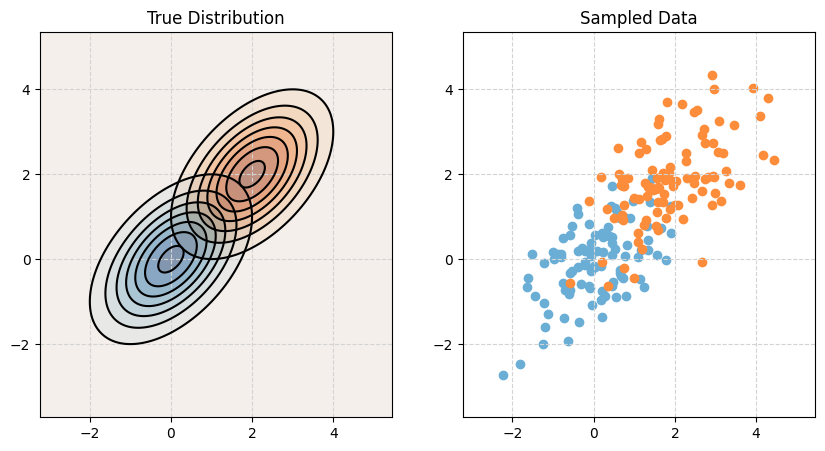
Quadratic Discriminant Analysis (QDA)
Quadratic Discriminant Analysis, QDA, also assumes that the features are multivariate normal conditioned on the target classes. However, unlike LDA, QDA makes fewer assumptions on the covariances (shapes). QDA allows the covariance to be completely different within each class.
\[ \boldsymbol X \mid Y = k \sim N(\boldsymbol\mu_k, \boldsymbol\Sigma_k) \]
# Make a two-class QDA setup
mu1, cov1 = create_params(0, 0, 2, 1, 0.5)
mu2, cov2 = create_params(2, 2, 1, 0.5, -0.5)
print_setup_parameter_info(mu1, cov1, mu2, cov2)Mean X when Y = Blue: [0 0]
Covariance of X when Y = Blue:
[[4. 1.]
[1. 1.]]
Mean X when Y = Orange: [2 2]
Covariance of X when Y = Orange:
[[ 1. -0.25]
[-0.25 0.25]]# Plot it!
make_plot(mu1, cov1, mu2, cov2)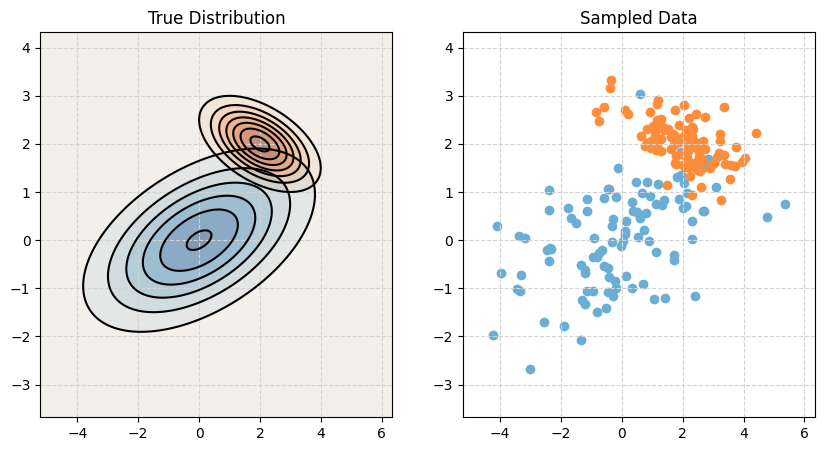
Naive Bayes (NB)
Naive Bayes comes in many forms. With only numeric features, it often assumes a multivariate normal conditioned on the target classes, but a very specific multivariate normal.
\[ {\boldsymbol X} \mid Y = k \sim N(\boldsymbol\mu_k, \boldsymbol\Sigma_k) \]
Naive Bayes assumes that the features \(X_1, X_2, \ldots, X_p\) are independent given \(Y = k\). This is the “naive” part of naive Bayes. The Bayes part is nothing new. Since \(X_1, X_2, \ldots, X_p\) are assumed independent, each \(\boldsymbol\Sigma_k\) is diagonal, that is, we assume no correlation between features. Independence implies zero correlation.
\[ \boldsymbol\Sigma_k = \begin{bmatrix} \sigma_{1}^2 & 0 & \cdots & 0 \\ 0 & \ddots & \ddots & \vdots \\ \vdots & \ddots & \ddots & 0 \\ 0 & \cdots & 0 & \sigma_{p}^2 \\ \end{bmatrix} \]
This will allow us to write the (joint) likelihood as a product of univariate distributions. In this case, the product of univariate normal distributions instead of a (joint) multivariate distribution.
\[ f_k(\boldsymbol x) = \prod_{j = 1}^{p} f_{kj}(\boldsymbol x_j) \]
Here, \(f_{kj}(\boldsymbol x_j)\) is the density for the \(j\)-th feature conditioned on the \(k\)-th class. Notice that there is a \(\sigma_{kj}\) for each feature for each class.
\[ f_{kj}(\boldsymbol x_j) = \frac{1}{\sigma_{kj}\sqrt{2\pi}}\exp\left[-\frac{1}{2}\left(\frac{x_j - \mu_{kj}}{\sigma_{kj}}\right)^2\right] \]
When \(p = 1\), this version of naive Bayes is equivalent to QDA.
# Make a two-class NB setup
mu1, cov1 = create_params(0, 0, 2, 1, 0)
mu2, cov2 = create_params(2, 2, 1, 0.5, 0)
print_setup_parameter_info(mu1, cov1, mu2, cov2)Mean X when Y = Blue: [0 0]
Covariance of X when Y = Blue:
[[4 0]
[0 1]]
Mean X when Y = Orange: [2 2]
Covariance of X when Y = Orange:
[[1. 0. ]
[0. 0.25]]# Plot it!
make_plot(mu1, cov1, mu2, cov2)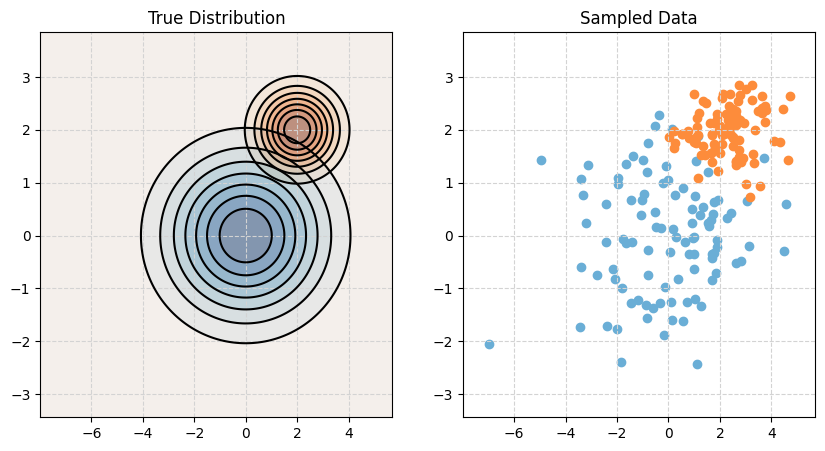
Generative Models with sklearn
# Function to generate data according to LDA, QDA, or NB
def generate_data(n1, n2, n3, mu1, mu2, mu3, cov1, cov2, cov3):
# Generate data for class A
data1 = np.random.multivariate_normal(mu1, cov1, n1)
labels1 = np.repeat("A", n1)
# Generate data for class B
data2 = np.random.multivariate_normal(mu2, cov2, n2)
labels2 = np.repeat("B", n2)
# Generate data for class C
data3 = np.random.multivariate_normal(mu3, cov3, n3)
labels3 = np.repeat("C", n3)
# Combine data and labels
data = np.concatenate((data1, data2, data3))
labels = np.concatenate((labels1, labels2, labels3))
# Make X and y
X = pd.DataFrame(data, columns=["x1", "x2"])
y = pd.Series(labels)
return X, y# Setup sample sizes
n1 = 500
n2 = 500
n3 = 500
# Setup parameters
mu1, cov1 = create_params(0, 0, 2, 1, 0.5)
mu2, cov2 = create_params(2, 2, 1, 2, -0.5)
mu3, cov3 = create_params(0, 2, 1, 1, 0.5)# Generate train and test data
X_train, y_train = generate_data(n1, n2, n3, mu1, mu2, mu3, cov1, cov2, cov3)
X_test, y_test = generate_data(n1, n2, n3, mu1, mu2, mu3, cov1, cov2, cov3)# Plot the train data
p = sns.jointplot(x="x1", y="x2", data=X_train, hue=y_train, space=0)
p.fig.suptitle("Simulated Train Data", y=1.01)
p.ax_joint.grid(True)
p.fig.set_size_inches(8, 8)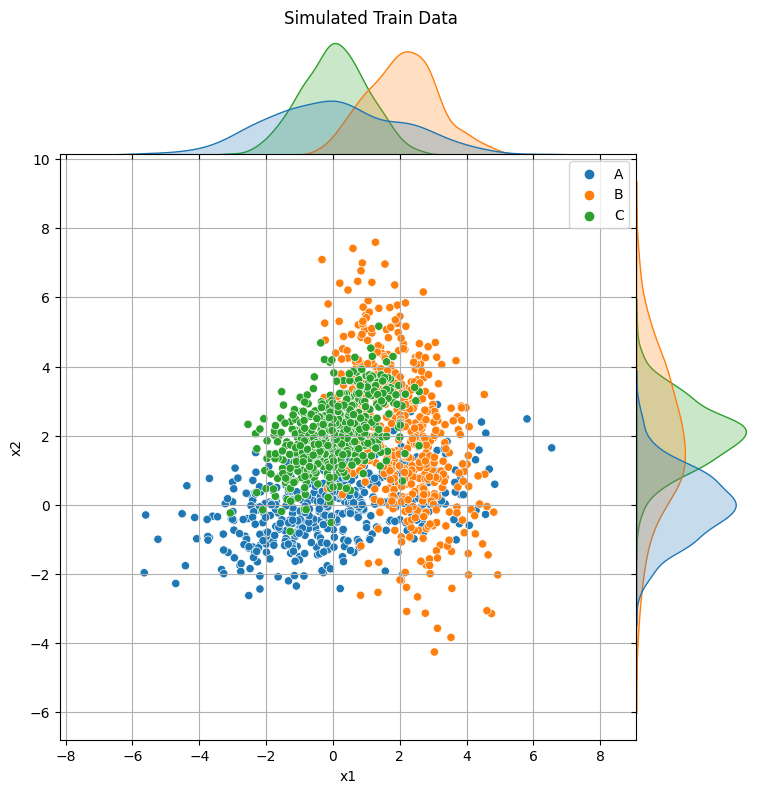
# Gaussian Naive Bayes
gnb = GaussianNB()
gnb.fit(X_train, y_train)
y_pred = gnb.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("Gaussian NB, Test Accuracy:", accuracy)
# Linear Discriminant Analysis
lda = LinearDiscriminantAnalysis(store_covariance=True)
lda.fit(X_train, y_train)
y_pred = lda.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("LDA, Test Accuracy:", accuracy)
# Quadratic Discriminant Analysis
qda = QuadraticDiscriminantAnalysis(store_covariance=True)
qda.fit(X_train, y_train)
y_pred = qda.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("QDA, Test Accuracy:", accuracy)Gaussian NB, Test Accuracy: 0.7466666666666667
LDA, Test Accuracy: 0.688
QDA, Test Accuracy: 0.758print("Classes:", gnb.classes_)
print("Feature names:", gnb.feature_names_in_)
print("NB, Estimated Class Priors:", gnb.class_prior_)
print("NB, Estimated Mean of each feature per class:", "\n", gnb.theta_)
print("NB, Estimated Variance of each feature per class:", "\n", gnb.var_)Classes: ['A' 'B' 'C']
Feature names: ['x1' 'x2']
NB, Estimated Class Priors: [0.33333333 0.33333333 0.33333333]
NB, Estimated Mean of each feature per class:
[[4.01566160e-02 2.38873059e-05]
[2.08143224e+00 1.97557520e+00]
[3.66427998e-02 2.06823633e+00]]
NB, Estimated Variance of each feature per class:
[[4.04651343 0.97403523]
[1.07528142 4.16455559]
[0.95628049 0.86020391]]print("Classes:", qda.classes_)
print("Feature names:", qda.feature_names_in_)
print("QDA, Estimated Class Priors:", qda.priors_)
print("QDA, Estimated Mean of each feature per class:", "\n", qda.means_)
print("QDA, Estimated Covariance Matrix of each feature per class:")
qda.covariance_Classes: ['A' 'B' 'C']
Feature names: ['x1' 'x2']
QDA, Estimated Class Priors: [0.33333333 0.33333333 0.33333333]
QDA, Estimated Mean of each feature per class:
[[4.01566160e-02 2.38873059e-05]
[2.08143224e+00 1.97557520e+00]
[3.66427998e-02 2.06823633e+00]]
QDA, Estimated Covariance Matrix of each feature per class:[array([[4.05462268, 0.92459548],
[0.92459548, 0.9759872 ]]),
array([[ 1.07743629, -0.94226649],
[-0.94226649, 4.17290139]]),
array([[0.95819688, 0.41460533],
[0.41460533, 0.86192776]])]Effect of Scaling
# Scale the X data
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)p = sns.jointplot(x="x1", y="x2", data=pd.DataFrame(X_train_scaled, columns=["x1", "x2"]), hue=y_train, space=0)
p.fig.suptitle("Simulated Train Data, Scaled", y=1.01)
p.ax_joint.grid(True)
p.fig.set_size_inches(8, 8)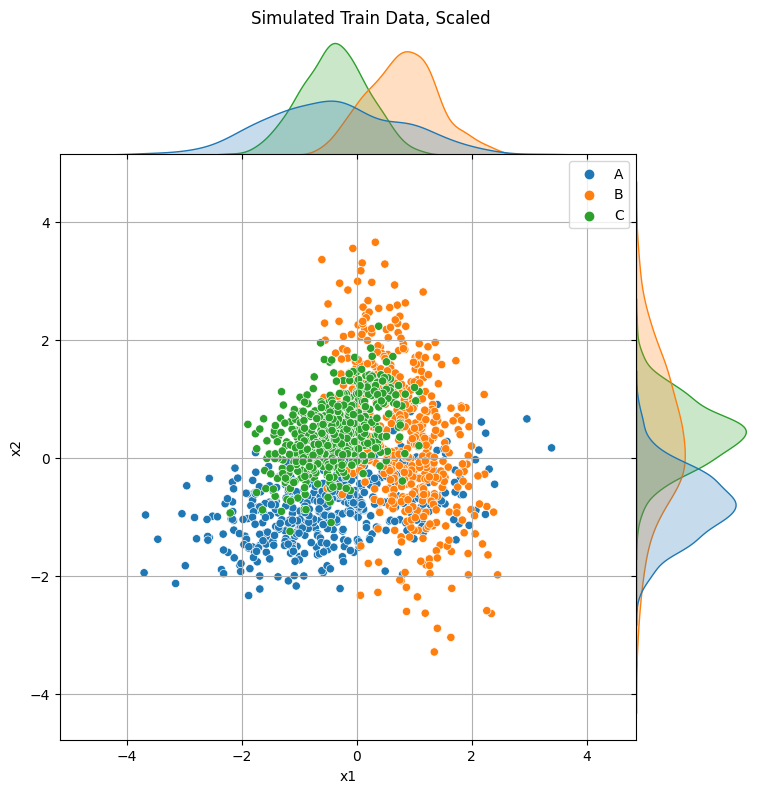
# Fit NB model again
gnbs = GaussianNB()
gnbs.fit(X_train_scaled, y_train)
y_pred = gnbs.predict(X_test_scaled)
# Calculate accuracy
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy)Accuracy: 0.7466666666666667# Check estimated probabilities
gnb.predict_proba(X_test) == gnbs.predict_proba(X_test_scaled)
np.allclose(gnb.predict_proba(X_test), gnbs.predict_proba(X_test_scaled))Trueprint("Classes:", gnbs.classes_)
print("NB, Estimated Class Priors:", gnbs.class_prior_)
print("NB, Estimated Mean of each feature per class:", "\n", gnbs.theta_)
print("NB, Estimated Variance of each feature per class:", "\n", gnbs.var_)Classes: ['A' 'B' 'C']
NB, Estimated Class Priors: [0.33333333 0.33333333 0.33333333]
NB, Estimated Mean of each feature per class:
[[-0.39523727 -0.79023671]
[ 0.79251912 0.36795645]
[-0.39728185 0.42228026]]
NB, Estimated Variance of each feature per class:
[[1.37003745 0.3347804 ]
[0.36406053 1.43137695]
[0.3237701 0.29565605]]Priors
# Setup sample sizes
n1_train = 500
n2_train = 300
n3_train = 700
n1_test = 500
n2_test = 500
n3_test = 500
# Setup parameters
mu1, cov1 = create_params(0, 0, 2, 1, 0.5)
mu2, cov2 = create_params(2, 2, 1, 2, -0.5)
mu3, cov3 = create_params(0, 2, 1, 1, 0.5)
# Generate train and test data
X_train, y_train = generate_data(n1_train, n2_train, n3_train, mu1, mu2, mu3, cov1, cov2, cov3)
X_test, y_test = generate_data(n1_test, n2_test, n3_test, mu1, mu2, mu3, cov1, cov2, cov3)# Quadratic Discriminant Analysis, Estimated Prior
qda = QuadraticDiscriminantAnalysis(store_covariance=True)
qda.fit(X_train, y_train)
y_pred = qda.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("QDA, Test Accuracy:", accuracy)QDA, Test Accuracy: 0.756# Quadratic Discriminant Analysis, Specified Prior
qda = QuadraticDiscriminantAnalysis(store_covariance=True, priors=[0.33, 0.33, 0.33])
qda.fit(X_train, y_train)
y_pred = qda.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("QDA, Test Accuracy:", accuracy)QDA, Test Accuracy: 0.764Penguins
from palmerpenguins import load_penguins
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.compose import ColumnTransformer
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV# Load the dataset
penguins = load_penguins()
# Convert to pandas dataframe
df = pd.DataFrame(penguins)
# Print the first 5 rows of the dataframe
df.head()| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | 2007 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | female | 2007 |
# Create X and y
df = df.drop("year", axis=1)
X = df.drop("species", axis=1)
y = df["species"]# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Print the shapes of the resulting dataframes
print("X_train shape:", X_train.shape)
print("y_train shape:", y_train.shape)
print("X_test shape:", X_test.shape)
print("y_test shape:", y_test.shape)X_train shape: (275, 6)
y_train shape: (275,)
X_test shape: (69, 6)
y_test shape: (69,)X_train| island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|
| 66 | Biscoe | 35.5 | 16.2 | 195.0 | 3350.0 | female |
| 229 | Biscoe | 51.1 | 16.3 | 220.0 | 6000.0 | male |
| 7 | Torgersen | 39.2 | 19.6 | 195.0 | 4675.0 | male |
| 140 | Dream | 40.2 | 17.1 | 193.0 | 3400.0 | female |
| 323 | Dream | 49.0 | 19.6 | 212.0 | 4300.0 | male |
| ... | ... | ... | ... | ... | ... | ... |
| 188 | Biscoe | 42.6 | 13.7 | 213.0 | 4950.0 | female |
| 71 | Torgersen | 39.7 | 18.4 | 190.0 | 3900.0 | male |
| 106 | Biscoe | 38.6 | 17.2 | 199.0 | 3750.0 | female |
| 270 | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | female |
| 102 | Biscoe | 37.7 | 16.0 | 183.0 | 3075.0 | female |
275 rows × 6 columns
y_train66 Adelie
229 Gentoo
7 Adelie
140 Adelie
323 Chinstrap
...
188 Gentoo
71 Adelie
106 Adelie
270 Gentoo
102 Adelie
Name: species, Length: 275, dtype: object# Define the numeric features
numeric_features = ["bill_length_mm", "bill_depth_mm", "flipper_length_mm", "body_mass_g"]
# Define the categorical features
categorical_features = ["sex", "island"]
# Define the column transformer
preprocessor = ColumnTransformer(
transformers=[
("num", StandardScaler(), numeric_features),
("cat", OneHotEncoder(), categorical_features),
]
)
# Define the full pipeline
pipe = Pipeline([("preprocessor", preprocessor), ("imputer", SimpleImputer()), ("classifier", SVC())])
# Define the parameter grid
param_grid = {
"imputer__strategy": ["mean", "median"],
"classifier__C": [0.1, 1, 10],
"classifier__kernel": ["linear", "rbf"],
}
# Define the grid search
grid = GridSearchCV(pipe, param_grid=param_grid, cv=5, verbose=1)
# Fit the grid search to the training data
grid.fit(X_train, y_train)
# Print the best parameters and score
print("Best parameters:", grid.best_params_)
print("Best score:", grid.best_score_)Fitting 5 folds for each of 12 candidates, totalling 60 fits
Best parameters: {'classifier__C': 0.1, 'classifier__kernel': 'linear', 'imputer__strategy': 'mean'}
Best score: 0.9927272727272728gridGridSearchCV(cv=5,
estimator=Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('num',
StandardScaler(),
['bill_length_mm',
'bill_depth_mm',
'flipper_length_mm',
'body_mass_g']),
('cat',
OneHotEncoder(),
['sex',
'island'])])),
('imputer', SimpleImputer()),
('classifier', SVC())]),
param_grid={'classifier__C': [0.1, 1, 10],
'classifier__kernel': ['linear', 'rbf'],
'imputer__strategy': ['mean', 'median']},
verbose=1)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GridSearchCV(cv=5,
estimator=Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('num',
StandardScaler(),
['bill_length_mm',
'bill_depth_mm',
'flipper_length_mm',
'body_mass_g']),
('cat',
OneHotEncoder(),
['sex',
'island'])])),
('imputer', SimpleImputer()),
('classifier', SVC())]),
param_grid={'classifier__C': [0.1, 1, 10],
'classifier__kernel': ['linear', 'rbf'],
'imputer__strategy': ['mean', 'median']},
verbose=1)Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('num', StandardScaler(),
['bill_length_mm',
'bill_depth_mm',
'flipper_length_mm',
'body_mass_g']),
('cat', OneHotEncoder(),
['sex', 'island'])])),
('imputer', SimpleImputer()), ('classifier', SVC())])ColumnTransformer(transformers=[('num', StandardScaler(),
['bill_length_mm', 'bill_depth_mm',
'flipper_length_mm', 'body_mass_g']),
('cat', OneHotEncoder(), ['sex', 'island'])])['bill_length_mm', 'bill_depth_mm', 'flipper_length_mm', 'body_mass_g']
StandardScaler()
['sex', 'island']
OneHotEncoder()
SimpleImputer()
SVC()
One-Hot Encoding
X_train_cat = X_train[categorical_features]
X_train_cat.to_numpy()[:10]array([['female', 'Biscoe'],
['male', 'Biscoe'],
['male', 'Torgersen'],
['female', 'Dream'],
['male', 'Dream'],
['male', 'Dream'],
['male', 'Dream'],
['female', 'Torgersen'],
['female', 'Dream'],
['female', 'Dream']], dtype=object)ohe = OneHotEncoder(sparse_output=False)
ohe.fit(X_train_cat)
ohe.fit_transform(X_train_cat)array([[1., 0., 0., 1., 0., 0.],
[0., 1., 0., 1., 0., 0.],
[0., 1., 0., 0., 0., 1.],
...,
[1., 0., 0., 1., 0., 0.],
[1., 0., 0., 1., 0., 0.],
[1., 0., 0., 1., 0., 0.]])de = OneHotEncoder(sparse_output=False, drop="first")
de.fit(X_train_cat)
de.fit_transform(X_train_cat)array([[0., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 1.],
...,
[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.]])Imputation
X_train_num = X_train[numeric_features]
X_train_num_nan = X_train_num[X_train.isna().any(axis=1)]
X_train_num_nan.to_numpy()array([[ 47.3, 13.8, 216. , 4725. ],
[ nan, nan, nan, nan],
[ 37.8, 17.1, 186. , 3300. ],
[ 44.5, 14.3, 216. , 4100. ],
[ 44.5, 15.7, 217. , 4875. ],
[ 46.2, 14.4, 214. , 4650. ],
[ 37.5, 18.9, 179. , 2975. ],
[ 37.8, 17.3, 180. , 3700. ],
[ 34.1, 18.1, 193. , 3475. ]])imp = SimpleImputer()
imp.fit(X_train_num)
imp.fit_transform(X_train_num_nan)array([[ 47.3 , 13.8 , 216. , 4725. ],
[ 41.2125, 16.2 , 200.125 , 3975. ],
[ 37.8 , 17.1 , 186. , 3300. ],
[ 44.5 , 14.3 , 216. , 4100. ],
[ 44.5 , 15.7 , 217. , 4875. ],
[ 46.2 , 14.4 , 214. , 4650. ],
[ 37.5 , 18.9 , 179. , 2975. ],
[ 37.8 , 17.3 , 180. , 3700. ],
[ 34.1 , 18.1 , 193. , 3475. ]])